pJAZZ-OC, pJAZZ-OK高难度基因克隆载体-BioVector NTCC质粒载体菌种细胞蛋白抗体基因保藏中心
- 价 格:¥17930
- 货 号:pJAZZ-OC, pJAZZ-OK
- 产 地:北京
- BioVector NTCC典型培养物保藏中心
- 联系人:Dr.Xu, Biovector NTCC Inc.
电话:400-800-2947 工作微信:1843439339 (QQ同号)
邮件:Biovector@163.com
手机:18901268599
地址:北京
- 已注册
pJAZZ-OC, pJAZZ-OK高难度基因克隆载体
描述:
用于易突变易重组基因的克隆,不易形成超螺旋结构减小扭力避免突变重组,提高基因稳定性。shotgun文库克隆理想载体。
BigEasy v2.0 Linear Cloning Kits (patent pending) provide an unprecedented ability to maintainDNAs that are otherwise unclonable. The BigEasy Kit is ideal for constructing shotgun libraries with largeinserts or for cloning smaller products, particularly when the target DNA is especially difficult to clone inconventional vectors.The BigEasy Kit is based on the novel linear cloning plasmids, pJAZZ® (Figure 1, Refs. 1-3), which arenot subject to supercoiling in the cell. Conventional circular plasmids are maintained in multiple states ofsupercoiling by the action of DNA topoisomerase and gyrase. Supercoiling induces torsional stress in theplasmid DNA, which is associated with structural instability of sequences that are AT-rich or containinverted repeats (4). The ends of the pJAZZ vectors can rotate freely as the molecule is replicated, so it is
not under torsional stress. As a result, numerous classes of structure-rich sequences are much morestable. In addition, the pJAZZ vectors incorporate Lucigen’s patented CloneSmart® technology fortranscription-free cloning (U.S. Pat. 6,709,861), which further reduces instability or loss of insert DNA.Large fragments or inserts with high AT content are cloned easily with this vector.The BigEasy Cloning Kits are convenient to use, containing pre-cut, dephosphorylated pJAZZ-OC orpJAZZ-OK cloning vector; DNATerminator End Repair enzymes and buffer (with the blunt kit only); ligaseand ligation buffer containing ATP; sequencing primers; competent cells; and DNA controls.Improvements over original BigEasy KitThe BigEasy v2.0 Kit contains two major changes from the original Big Easy Kit (see Table below). Thenew pJAZZ-OC vector is resistant only to chloramphenicol, and the new pJAZZ-OK vector is resistant toonly kanamycin. The original pJAZZ-KA vector was resistant to kanamycin plus ampicillin.The new BigEasy TSA™ Electrocompetent Cells have an ampicillin resistance gene integrated into thechromosome and no exogenous plasmids. The original BigEasy pTel cells had an integratedchloramphenicol gene and an exogenous plasmid that encoded gentamycin resistance.Kit Version Vector CellsBigEasy v2.0 pJAZZ-OC (chloramphenicolR)pJAZZ-OK (kanamycinR)BigEasy TSA: ampicillinRNo Endogenous plasmidsOriginal BigEasy pJAZZ-KAkanamycinR + ampicillinRBigEasy pTel: chloramphenicolRgentamycinR on endogenous plasmidpJAZZ®-OC and pJAZZ-OK VectorsThe pJAZZ-OC and –OK vectors are supplied pre-digested at SmaI (blunt) or NotI sites, and they havedephosphorylated ends. Each of the vectors contains a pair of nearly identical Multiple Cloning Sites(Figure 1). During preparation of the vector, both Multiple Cloning Sites are cleaved by restrictiondigestion, then dephosphorylated, preventing their re-ligation. Insert DNA is ligated between the two armsto re-create a viable linear plasmid.
Map图谱:
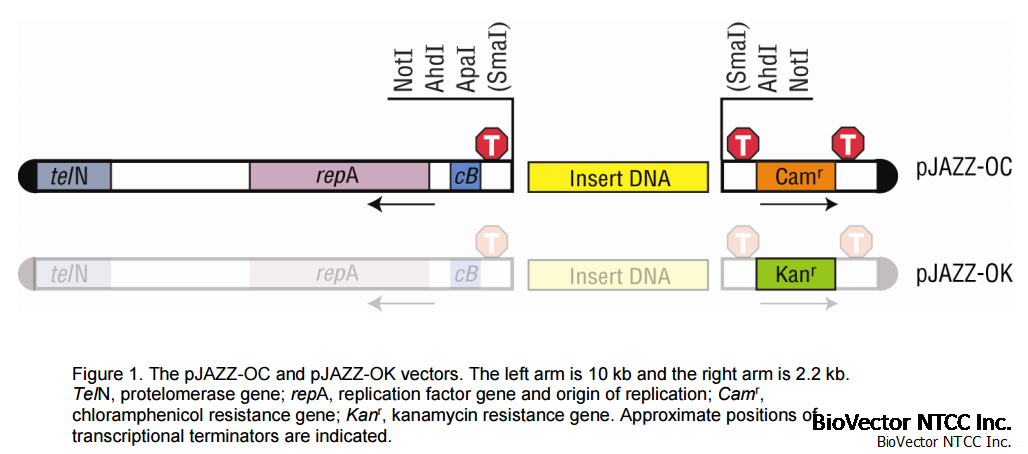
Figure 1. The pJAZZ-OC and pJAZZ-OK vectors. The left arm is 10 kb and the right arm is 2.2 kb.TelN, protelomerase gene; repA, replication factor gene and origin of replication; Camr,chloramphenicol resistance gene; Kanr, kanamycin resistance gene. Approximate positions oftranscriptional terminators are indicated.
The pJAZZ-OC and -OK vectors also employ the CloneSmart transcription-free cloning technology (USPatent #6,709,861), which eliminates transcription both into and out of the insert DNA. Thus, clonedfragments are not subjected to vector-driven transcription. In conventional plasmids, inserts are cloneddownstream of a strong promoter, typically within the coding sequence of lacZ or a negative selectiongene, such as ccdB. Transcription from the promoter causes loss of plasmids containing toxic codingsequences, strong secondary structure, or other deleterious features. In the pJAZZ vectors (as in mostLucigen vectors), transcription across the insert is avoided, so this loss is minimized.Inserts containing E. coli-like promoters are often difficult to clone in conventional plasmids, becausetranscription from these promoters can interfere with the plasmid’s replication or expression of its drugresistance gene. In Lucigen’s pJAZZ vectors, strong transcription terminators flank the cloning site toblock this transcription, eliminating another source of cloning bias and sequencing gaps.The left arm of the vector contains the origin of replication, and the right arm encodes resistance tochloramphenicol or kanamycin (Figure 1). Selection with the appropriate antibiotic results in recombinantclones containing both vector arms flanking the insert DNA.The pJAZZ-OC and -OK vectors contain an inducible origin of replication. The copy number is ~2-4/cellprior to induction; it is increased by approximately 5-20 fold by induction in BigEasy TSA cells (seebelow).The GenBank Accession number of the pJAZZ-OC vector is EF583812; the Accession number for thepJAZZ-OK vector is FJ160465.BigEasy TSA™ Electrocompetent CellsThe BigEasy TSA strain is derived from Lucigen’s E. cloni® 10G cells. This strain contains several genesfrom phage N15, which are essential for high transformation efficiency and induction of copy number ofthe pJAZZ linear vectors. Transformation with other strains will be 20-200 X less efficient. The N15genes are telN, which encodes pro-telomerase for efficient replication of the linear plasmid; the sopABgenes for stable inheritance; and the antA gene to regulate copy number.These cells give high yield and high quality plasmid DNA due to the endA1 and recA1 mutations. Theycontain the mcr and mrr mutations, which allow stable cloning of methylated genomic DNA that has beenisolated directly from mammalian or plant cells. The rpsL mutation confers resistance to streptomycin.BigEasy TSA cells contain an integrated copy of the bla gene; they are therefore resistant to ampicillin.BigEasy TSA Genotype:F- mcrA (mrr-hsdRMS-mcrBC) 80dlacZM15 lacX74 endA1 recA1araD139 (ara, leu)7697galU galK rpsL (StrR) nupG - tonA bla (AmpR) sopAB telN antA BigEasy TSA Electrocompetent Cells are provided with supercoiled control pKanR plasmid DNA at aconcentration of 10 pg/µL. The pKanR control plasmid is kanamycin resistant.For highest transformation efficiency, use the provided Recovery Medium to resuspend the cellsafter electroporation. Use of TB, SOC, or other media may result in lower transformationefficiencies.Purification and Size Fractionation of DNADNA must be purified from restriction or repair enzymes before ligation to pJAZZ vectors. Agarose gelelectrophoresis, which is commonly used to size fractionate DNA fragments, is sufficient for purification. Ifthe insert DNA is not fractionated by electrophoresis after repair or digestion, it must be purified bybinding to a DNA purification column or by phenol/chloroform extraction to remove the repair enzymes.
Sensitivity of DNA to Short Wavelength UV Light
DNA resolved on agarose gels is generally stained with ethidium bromide and visualized by illuminationwith ultraviolet light. Exposure to short wavelength ultraviolet light (e.g., 254, 302, or 312 nm) can reducecloning efficiencies by several orders of magnitude (Figure 2). Note that the wavelength of most UVtransilluminators, even those designated specifically for DNA visualization, is typically 302 nm or 312 nm,and can cause significant damage to DNA.No UV 30s302nm60s302nm120s302nm120s360nmFigure 2. Relative cloning efficiency of pUC19 after exposure to short or long wavelength UV light. IntactpUC19 DNA was transformed after no UV exposure (“No UV”) or exposure to 302 nm UV light for 30, 60,or 90 seconds (“30 s 30 2nm, 60s 302 nm,120 s 302 nm”) or to 360 nm UV light for 120 seconds (“120 s360 nm”). Cloning efficiencies were calculated relative to un-irradiated pUC19 DNA.To safely gel-isolate large fragments, we recommend running a duplicate lane of the insert DNA. Stain,photograph, and physically mark the band in the duplicate lane, place it next to an unstained lanecontaining the desired sample, and excise the region of the unmarked gel corresponding to the position ofthe desired band. This method completely avoids exposure of the band to ethidium bromide and UV light.
BioVector NTCC质粒载体菌种细胞蛋白抗体基因保藏中心提供
- 公告/新闻